PROJECT
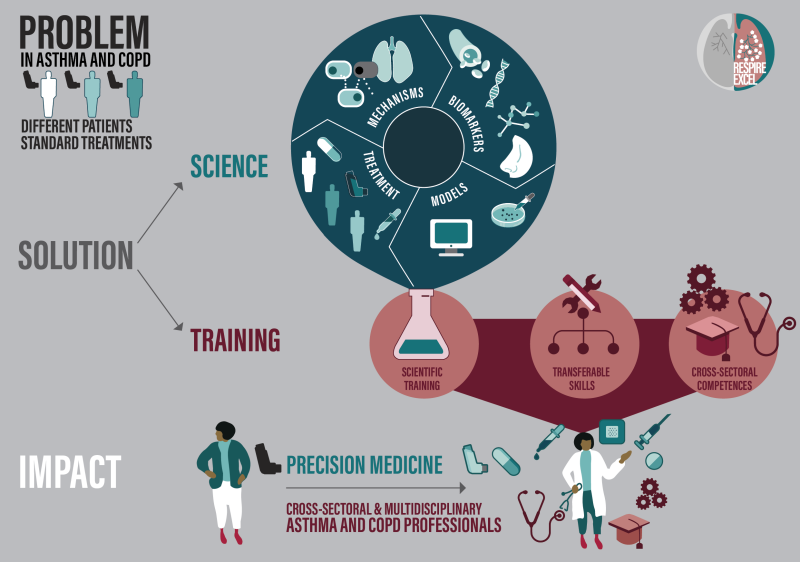
Asthma and COPD are common chronic respiratory diseases that represent a significant social and economic burden. Traditionally, treatment of these conditions has been based on a "one-size-fits-all" approach, which often suppresses symptoms without achieving actual health improvement. RESPIRE-EXCEL aims to change this by introducing precision medicine tailored to individual patients. The project trains a new generation of translational and medical scientists in cross-sectoral competencies and multidisciplinary knowledge, so that they can effectively collaborate with industry, healthcare and academia to develop precision medicine for asthma and COPD. The RESPIRE-EXCEL consortium has many academic, industrial and societal partners from all over Europe, including the patient organization European Lung Foundation and the largest European scientific and clinical organization in respiratory medicine, the European Respiratory Society.
The objectives of the network are as follows:
- Identify molecular mechanisms – RESPIRE-EXCEL will unravel the complex molecular underlying mechanisms of asthma and COPD, enabling targeted interventions.
- Develop diagnostic biomarkers – RESPIRE-EXCEL will create improved prognostic and diagnostic biomarkers, enabling early detection of disease activity in individual patients.
- Support new treatments – RESPIRE-EXCEL will advance the development of precision medicine, targeting specific disease mechanisms to ultimately achieve stable health improvement in patients.
The RESPIRE-EXCEL project illustrates the power of interdisciplinary collaboration and highlights the essential role of precision medicine in improving health outcomes for patients with asthma and COPD.
WORK PACKAGES
WP2 | Charting Cellular & Molecular Mechanisms of Disease (Mechanisms)
To define endotypes of asthma and COPD, we need to understand the cellular and molecular mechanisms in the airway wall and lung tissue. We will map the gene regulatory networks (GRNs) in lung tissue using existing datasets from UMCG, HMGU, IPMC, and the literature. Advanced techniques like machine learning and generative models will help predict responses that can be tested in model systems. We will use spatial transcriptomics, proteomics, and lipidomics to study lung tissue from patients with asthma and COPD. Spatial transcriptomics will be performed on bronchial biopsies and lung tissue samples, complemented by targeted imaging of lipid mediators and spatial proteomic analyses. These comprehensive analyses will generate detailed datasets, allowing us to identify multiple causative mechanisms of disease and potential treatment targets. The integrated analyses will be supported by RESPIRE-EXCEL partners, ensuring a thorough understanding of the disease mechanisms.
WP3 | Identifying Disease Endotypes for Patient Stratification (Biomarkers)
To stratify patients for precision medicine, we need biomarkers in nasal swabs or blood that reflect disease mechanisms in lung tissue with high sensitivity and specificity. We will analyze samples from asthma and COPD patients, covering various disease stages, using multimodal techniques like RNA-Seq, DNA-methyl Seq, and high-dimensional flow cytometry. This will help identify stable patient clusters and map them to cellular mechanisms of disease. We will perform in-depth analyses of the airway epithelium to understand how viral infections lead to airway remodeling in asthma. In COPD, we will conduct longitudinal analyses of the airway wall transcriptome and methylome in matched bronchial and nasal samples. Additionally, we will study changes in the adaptive immune receptor repertoire during disease onset, progression, and exacerbations. These comprehensive analyses will help distinguish endotypes of asthma and COPD, aiding in patient stratification for targeted treatments.
WP4 | Cellular & Molecular Basis for Treatment Responses to Biologicals (Treatment)
To achieve disease remission with precision medicine, we need to understand which endotypes of asthma and COPD respond to treatments. We will study treatment responses in patients using biologicals like Tezepelumab and Dupilumab, analyzing samples before and after treatment. This will involve performing scRNA-seq and highly multiplexed immunohistochemical analyses on airway wall biopsies, complemented by functional studies in cultured bronchial epithelial cells. We will also analyze treatment response to Mepolizumab in patients with severe asthma, using scRNA-seq, spatial transcriptomics, and high-dimensional flow cytometry. Additionally, we will compare immune cell subsets in blood and bronchoalveolar lavage in severe COPD patients before and after treatment with Tezepelumab. These detailed analyses will help identify responder and non-responder endotypes, explore new treatment options, and understand the cellular and molecular basis for treatment responses.
WP5 | Endotype-based Models for Validation of Targets and Biomarkers (Models)
Current treatments may not cover all endotypes of asthma and COPD. We will identify and validate targets for additional endotypes. RESPIRE-EXCEL will create a comprehensive disease atlas of the human lung, integrating various datasets from asthma and COPD patients. We will also develop COPD model atlases, incorporating scRNA-seq data from patients, cultured primary cells, and mouse models. This multi-species COPD model atlas will guide the selection of optimal disease models for target validation and drug discovery. Additionally, we will generate primary cell culture models reflecting endotype-specific cellular mechanisms. Using air-liquid interface and 3D epithelial organoid culture models, we will study asthma-specific responses to various stimuli and test new treatments.